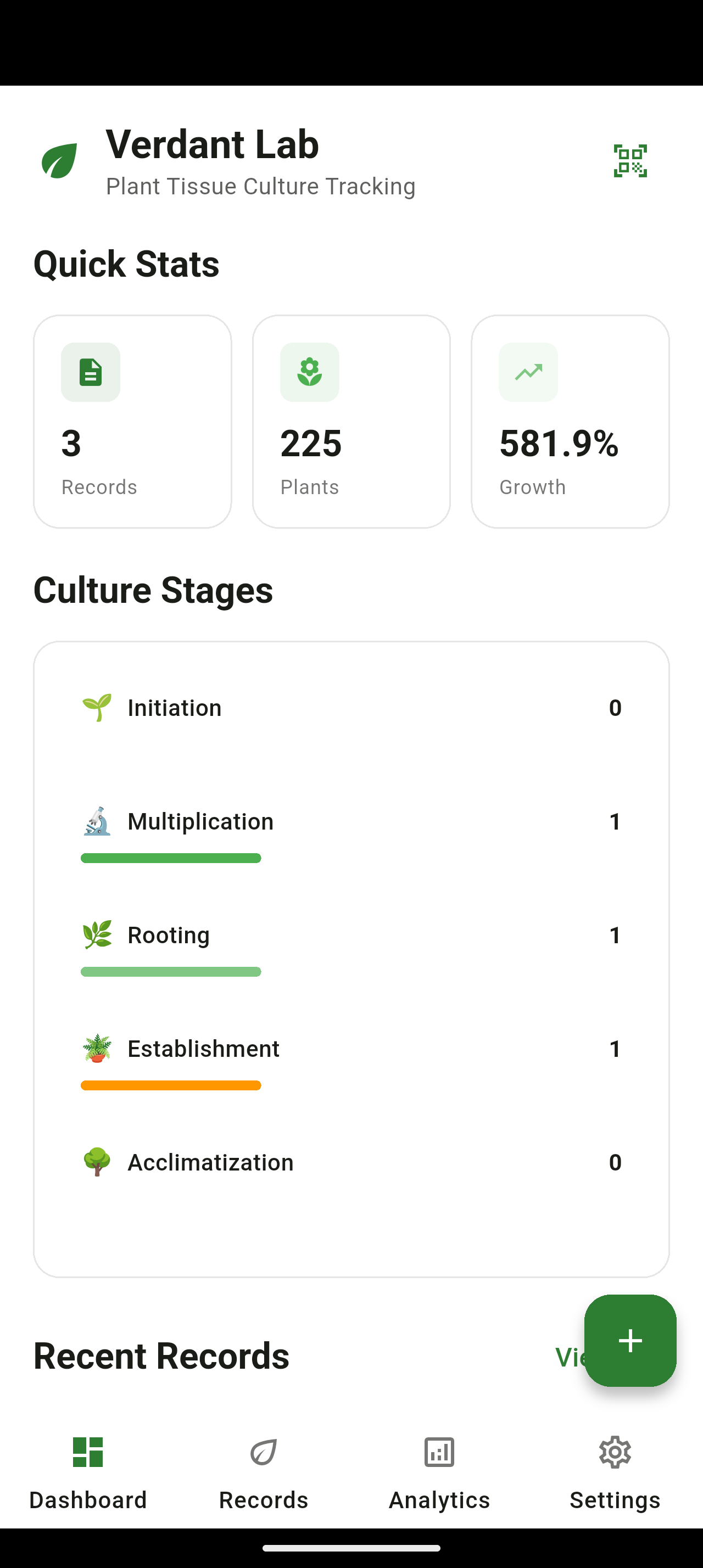
Dashboard featuring hierarchical lineage tracking, contamination alerts, and active culture management.
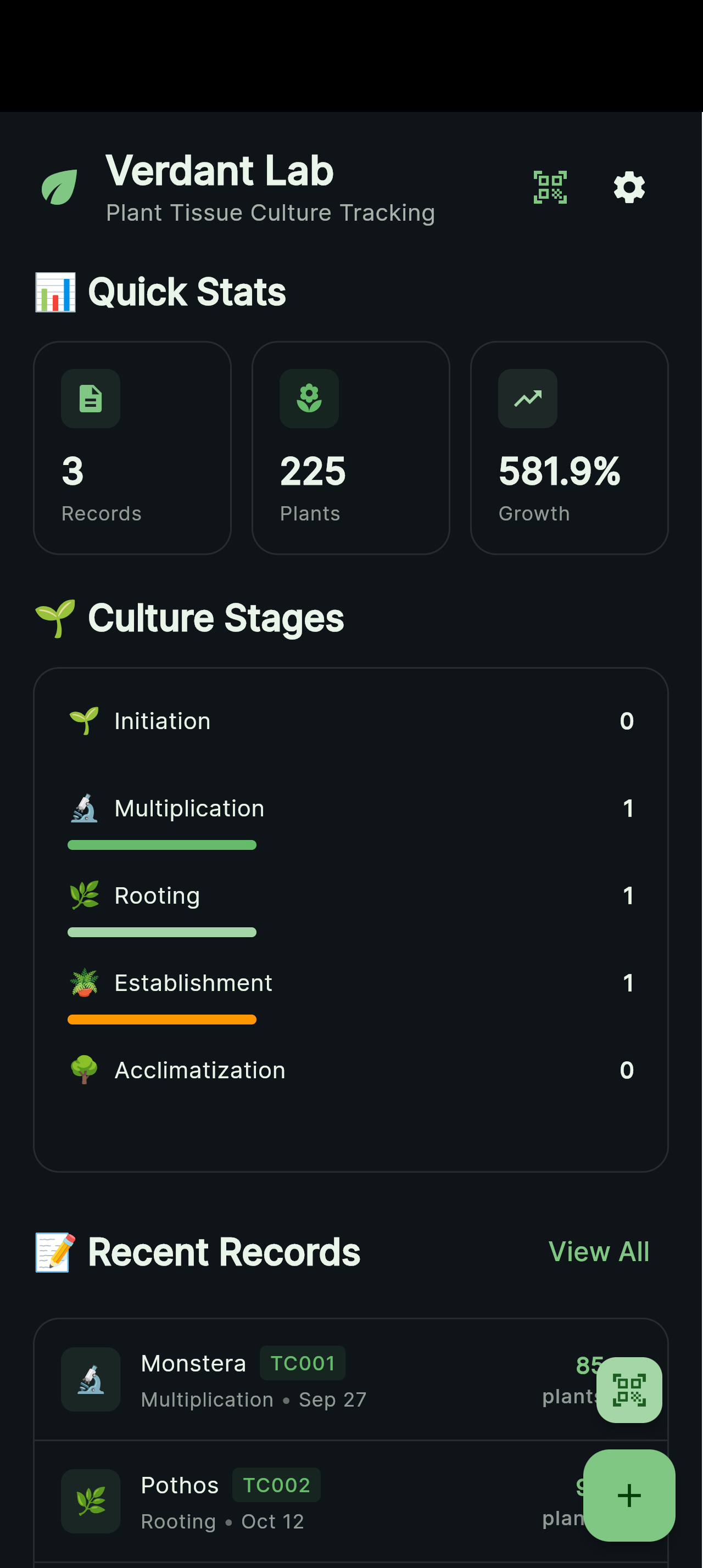
Home screen with active cultures, recent activity, and quick access to lineage trees.
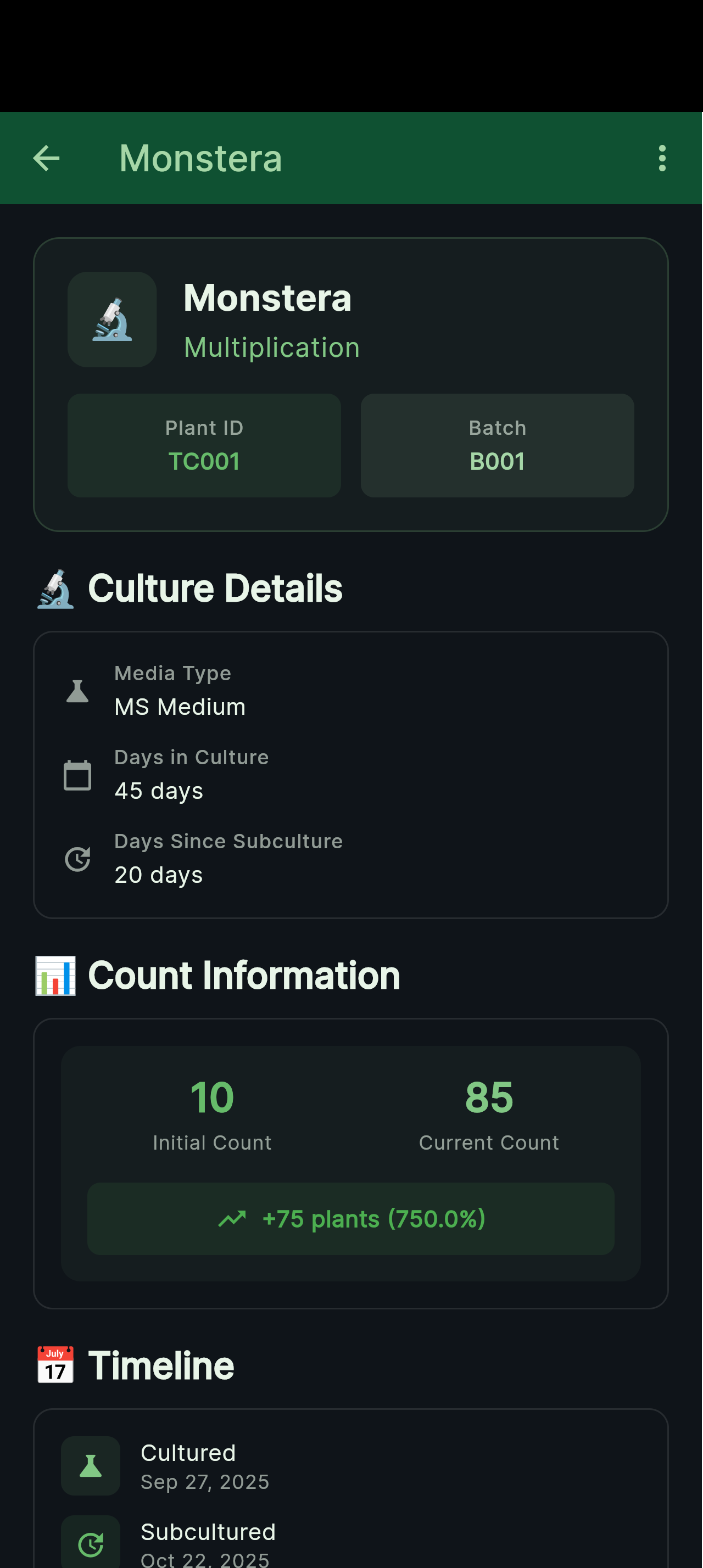
Culture detail view showing parent lineage, medium formulation, and contamination status.
5-Level Hierarchical Tracking
Recursive parent-child relationships for propagation lineages from mother plant through sub-subcultures.
Medium Formulations
Precise nutrient recipes with growth regulator concentrations and pH targets for reproducible results.
Contamination Analytics
Pattern identification by species, medium, and source material to reduce loss rates over time.
Verdant Lab designed for mobile-first tissue culture management in the lab and greenhouse.
Account management with profile details and data export options.
Clean login experience with secure authentication flow.
Built on Proven Architecture
Verdant Lab was adapted from HydroMate's reusable component architecture in just 3 days, demonstrating the power of designing for extensibility from the start.
Printory
Core architecture — 2 months
HydroMate
Adapted in 3 days
Verdant Lab
Adapted in 3 days
Settings and configuration for culture tracking preferences, notification thresholds, and data management.
The Process
Tissue culture propagation involves multi-stage tracking with deeply hierarchical data — a mother plant produces explants, which produce subcultures, which produce further divisions across different growth media. Researchers tracking this in lab notebooks lose visibility into contamination patterns and lineage history. Enterprise LIMS solutions exist but start at $10k+, putting them out of reach for independent researchers and small-scale propagators.
Building on HydroMate's proven architecture, I designed recursive data structures capable of representing 5+ levels of lineage tracking. The core challenge was mobile-optimized visualization of tree structures — making deeply nested parent-child relationships navigable on a phone screen without losing context. Medium formulation tracking required precise numeric inputs for nutrient concentrations and growth regulators while remaining fast enough for lab use with gloved hands.
Closed testing with 3 users validated the approach immediately. The contamination analytics feature revealed patterns that were never visible in paper notebooks — correlations between specific media formulations, source species, and contamination rates that allowed users to adjust protocols proactively. This confirmed that reusable architectures designed for inventory tracking can successfully extend into complex biological research domains. Validating that the interaction language translates across domains--from hydroponic monitoring to tissue culture tracking--confirmed our recursive data model approach.
PLACEHOLDER: Recursive lineage tree diagram showing mother plant through sub-subcultures
Hierarchical lineage tracking
PLACEHOLDER: Medium formulation database showing nutrient concentrations and pH targets
Medium formulation database
PLACEHOLDER: Contamination rate analytics dashboard with pattern identification
Contamination analytics dashboard
Impact
Concept to Launch
Closed Testing Users
Hierarchical Data Depth